Evaluation of Type 2 Diabetes Risk Variants (Alleles) in the Pashtun Ethnic Population of Pakistan
DOI:
https://doi.org/10.15605/jafes.037.S3Keywords:
type 2 diabetes, risk variants, bioinformatics, whole exome sequencing, Pashtun populationAbstract
Objective. To evaluate the Type 2 Diabetes (T2D) risk variants in the Pashtun ethnic population of Khyber Pakhtunkhwa using nascent whole-exome sequencing (WES) to better understand the pathogenesis of this complex polygenic disorder.
Methodology. A total of 100 confirmed patients with T2D of Pashtun ethnicity were included in the study, DNA was extracted from whole blood samples, and paired-end libraries were prepared using the Illumina Nextera XT DNA library kit carefully following the manufacturer’s instructions. Illumina HiSeq 2000 was used to obtain sequences of the prepared libraries followed by bioinformatics data analysis.
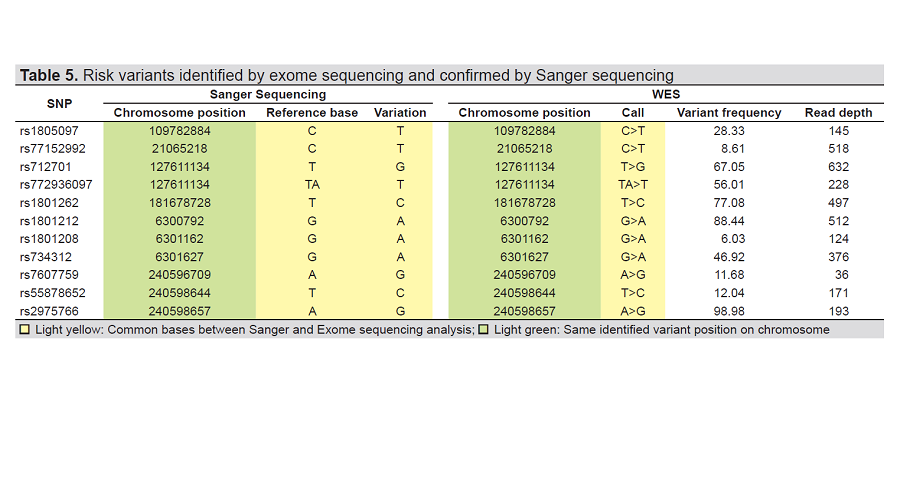
Results. A total of n=11 pathogenic/likely pathogenic variants were reported in the CAP10, PAX4, IRS-2, NEUROD1, CDKL1 and WFS1. Among the reported variants CAP10/rs55878652 (c.1990-7T>C; p.Leu446Pro) and CAP10/rs2975766 (c.1996A>G; p.Ile666Val) identified were novel, and have not yet been reported for any disease in the database. The variants CAP10/rs7607759 (c.1510A>G, p.Thr504Ala), PAX4/rs712701 (c.962A>C; p.His321Pro), PAX4/ rs772936097 (c.748-3delT; p.Arg325Trp), IRS-2/rs1805097 (c.3170G>A; p.Gly1057Asp), NEUROD1/rs1801262 (c.133A>G; p.Thr45Ala), CDKL1/rs77152992 (c.1226C>T; p.Pro409Leu), WFS1/rs1801212 (c.997G>A; p.Val333Ile), WFS1/rs1801208 (c.1367G>A; p.Arg456His), and WFS1/rs734312 (c.1832G>A; p.Arg611His) are previously identified in other ethnic populations. Our study reconfirms the associations of these variants with T2D in the Pakistani Pashtun population.
Conclusion. In-silico analysis of exome sequencing data suggests a statistically substantial association of all (n=11)
identified variants with T2D in the Pashtun ethnic population. This study may serve as a foundation for performing
future molecular studies aimed at unraveling T2D associated genes.
Downloads
References
Adeghate E, Schattner P, Dunn E. An update on the etiology and epidemiology of diabetes mellitus. Ann N Y Acad Sci. 2006;1084:1-29. https://pubmed.ncbi.nlm.nih.gov/17151290. https://doi.org/10.1196/annals.1372.029.
Mayor S. Diabetes affects nearly 6% of the world's adults. BMJ. 2006;333(7580):1191. https://pubmed.ncbi.nlm.nih.gov/7158376. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1693646. https://doi.org/10.1136/bmj.39055.608507.DB.
Boutayeb A. The double burden of communicable and non-communicable diseases in developing countries. Trans R Soc Trop Med Hyg. 2006;100(3):191-9. https://pubmed.ncbi.nlm.nih.gov/16274715. https://doi.org/10.1016/j.trstmh.2005.07.021.
Al-Mawali A. Non-communicable diseases: Shining a light on cardiovascular disease, Oman’s biggest killer. Oman Med J. 2015;30(4):227. https://pubmed.ncbi.nlm.nih.gov/26366254. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4561645. https://doi.org/10.5001/omj.2015.47.
Saeedi P, Petersohn I, Salpea P, et al. Global and regional diabetes prevalence estimates for 2019 and projections for 2030 and 2045: Results from the International Diabetes Federation Diabetes Atlas. Diabetes research and clinical practice. 2019 ; 11(157):107843. https://pubmed.ncbi.nlm.nih.gov/31518657. https://doi.org/10.1016/j.diabres.2019.107843
Basit A, Fawwad A, Qureshi H, Shera A. Prevalence of diabetes, pre-diabetes and associated risk factors: Second National Diabetes Survey of Pakistan (NDSP), 2016–2017. BMJ Open. 2018;8(8):e020961. https://pubmed.ncbi.nlm.nih.gov/30082350. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6078264. https://doi.org/10.1136/bmjopen-2017-020961.
Hills AP, Misra A, Gill JM, et al. Public health and health systems: Implications for the prevention and management of type 2 diabetes in south Asia. Lancet Diabetes Endocrinol. 2018;6(12):992-1002. https://pubmed.ncbi.nlm.nih.gov/30287104. https://doi.org/10.1016/S2213-8587(18)30203-1.
Ghaffar A, Reddy KS, Singhi M. Burden of non-communicable diseases in South Asia. BMJ. 2004;328(7443):807-10. https://pubmed.ncbi.nlm.nih.gov/15070638. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC383378. https://doi.org/10.1136/bmj.328.7443.807.
Garduño-Diaz SD, Khokhar S. Prevalence, risk factors and complications associated with type 2 diabetes in South Asian migrants. Diabetes Metab Res Rev. 2012;28(1):6-24. https://pubmed.ncbi.nlm.nih.gov/21591242. https://doi.org/10.1002/dmrr.1219.
Adnan M, Aasim M. Prevalence of type 2 diabetes mellitus in the adult population of Pakistan: A meta-analysis of prospective cross-sectional surveys. Ann Glob Health. 2020;86(1):7. https://pubmed.ncbi.nlm.nih.gov/32025503. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6993597. https://doi.org/10.5334/aogh.2679.
Jabbar A, Farooqui K, Habib A, et al. Clinical characteristics and outcomes of diabetic ketoacidosis in Pakistani adults with type 2 diabetes mellitus. Diabet Med. 2004;21(8):920-3. https://pubmed.ncbi.nlm.nih.gov/15270798. https://doi.org/10.1111/j.1464-5491.2004.01249.x.
Jafar T, Levey A, White F, et al. Ethnic differences and determinants of diabetes and central obesity among South Asians in Pakistan. Diab Med. 2004;21(7):716-23. https://pubmed.ncbi.nlm.nih.gov/15209764. https://doi.org/10.1111/j.1464-5491.2004.01140.x.
Rees SD, Hydrie MZI, Shera AS, et al. Replication of 13 genome-wide association (GWA)-validated risk variants for type 2 diabetes in Pakistani populations. Diabetologia. 2011;54(6):1368-74. https://pubmed.ncbi.nlm.nih.gov/21350842. https://doi.org/ 10.1007/s00125-011-2063-2.
Zeggini E, McCarthy MI. TCF7L2: The biggest story in diabetes genetics since HLA? Diabetologia. 2007;50(1):1-4. https://pubmed.ncbi.nlm.nih.gov/17096114. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1760757. https://doi.org/10.1007/s00125-006-0507-x.
Saxena R, Elbers CC, Guo Y, et al. Large-scale gene-centric meta-analysis across 39 studies identifies type 2 diabetes loci. Am J Hum Genet. 2012;90(3):410-25. https://pubmed.ncbi.nlm.nih.gov/22325160. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3309185. https://doi.org/10.1016/j.ajhg.2011.12.022.
van Tilburg J, van Haeften TW, Pearson P, Wijmenga C. Defining the genetic contribution of type 2 diabetes mellitus. J Med Genet. 2001;38(9):569-78. https://pubmed.ncbi.nlm.nih.gov/11546824. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1734947. https://doi.org/10.1136/jmg.38.9.569.
Helgason A, Pálsson S, Thorleifsson G, et al. Refine the impact of variants of the TCF7L2 gene on type 2 diabetes and adaptive evolution. Nat Genet. 2007;39(2):218-25. https://pubmed.ncbi.nlm.nih.gov/17206141.
Sesti G, Federici M, Hribal ML, Lauro D, Sbraccia P, Lauro R. Defects of the insulin receptor substrate (IRS) system in human metabolic disorders. FASEB J. 2001;15(12):2099-111. https://pubmed.ncbi.nlm.nih.gov/11641236. https://doi.org/10.1096/fj.01-0009rev.
Liu Y, Yu L, Zhang D, et al. Positive association between variations in CDKAL1 and type 2 diabetes in Han Chinese individuals. Diabetologia. 2008;51(11):2134-7. https://pubmed.ncbi.nlm.nih.gov/18766326. https://doi.org/10.1007/s00125-008-1141-6.
Wang H, Rissanen J, Miettinen R, et al. New amino acid substitutions in the IRS-2 gene in Finnish and Chinese subjects with late-onset type 2 diabetes. Diabetes. 2001;50(8):1949-51. https://pubmed.ncbi.nlm.nih.gov/11473060. https://doi.org/10.2337/diabetes.50.8.1949.
Kanatsuka A, Tokuyama Y, Nozaki O, Matsui K, Egashira T. Beta-cell dysfunction in late-onset diabetic subjects carrying homozygous mutation in transcription factors NeuroD1 and Pax4. Metabolism. 2002;51(9):1161-5. https://pubmed.ncbi.nlm.nih.gov/12200761. https://doi.org/ 10.1053/meta.2002.34707.
Bonnefond A, Froguel P. Rare and common genetic events in type 2 diabetes: What should biologists know? Cell Metabol. 2015;21(3):357-68. https://pubmed.ncbi.nlm.nih.gov/25640731. https://doi.org/10.1016/j.cmet.2014.12.020.
Purcell S, Cherny SS, Sham PC. Genetic power calculator: Design of linkage and association genetic mapping studies of complex traits. Bioinformatics. 2003;19(1):149-50. https://pubmed.ncbi.nlm.nih.gov/12499305. https://doi.org/10.1093/bioinformatics/19.1.149.
Wang X, Sui W, Wu W, et al. Whole genome resequencing of 100 healthy individuals using DNA pooling. Exp Ther Med. 2016;12(5):3143-50. https://pubmed.ncbi.nlm.nih.gov/27882129. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5103757. https://doi.org/10.3892/etm.2016.3797.
Amos CI, Frazier ML, Wang W. DNA pooling in mutation detection with reference to sequence analysis. Am J Hum Genet. 2000;66(5):1689-92. https://pubmed.ncbi.nlm.nih.gov/10733464. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1378002. https://doi.org/10.1086/302894.
Carmi R, Rokhlina T, Kwitek-Black AE, et al. Use of a DNA pooling strategy to identify a human obesity syndrome locus on chromosome 15. Human molecular genetics. 1995;4(1):9-13. https://pubmed.ncbi.nlm.nih.gov/7711739. https://doi.org/10.1093/hmg/4.1.9.
Druley TE, Vallania FLM, Wegner DJ, et al. Quantification of rare allelic variants from pooled genomic DNA. Nat Methods. 2009;6(4):263-5. https://pubmed.ncbi.nlm.nih.gov/19252504. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2776647. https://doi.org/10.1038/nmeth.1307.
Hess J, Kohl T, Kotrová M, et al. Library preparation for next generation sequencing: A review of automation strategies. Biotechnol Adv. 2020;41:107537. https://pubmed.ncbi.nlm.nih.gov/32199980. https://doi.org/10.1016/j.biotechadv.2020.107537.
Nextera® XT DNA sample preparation guide. Illumina Inc.; 2012. https://hahana.soest.hawaii.edu/cmoreserver/summercourse/2015/documents/Metagenomics_06-22/nextera_xt_sample_preparation_guide_15031942_c.pdf. Accessed December 22, 2020.
Marine R, Polson SW, Ravel J, et al. Evaluation of a transposase protocol for rapid generation of shotgun high-throughput sequencing libraries from nanogram quantities of DNA. Appl Environ Microbiol. 2011;77(22):8071-9. https://pubmed.ncbi.nlm.nih.gov/21948828. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3209006. https://doi.org/10.1128/AEM.05610-11.
Adey A, Morrison HG, Xun X, et al. Rapid, low-input, low-bias construction of shotgun fragment libraries by high-density in vitro transposition. Genome Biol. 2010;11(12):R119. https://pubmed.ncbi.nlm.nih.gov/21143862. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3046479. https://doi.org/10.1186/gb-2010-11-12-r119.
Bronner IF, Quail MA, Turner DJ, Swerdlow H. Improved protocols for illumina sequencing. Curr Protoc Hum Genet. 2014;80:18.2.1-42. https://pubmed.ncbi.nlm.nih.gov/26270174. https://doi.org/10.1002/0471142905.hg1802s80.
Jones MR, Good JM. Targeted capture in evolutionary and ecological genomics. Mol Ecol. 2016;25(1):185-202. https://pubmed.ncbi.nlm.nih.gov/26137993. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4823023. https://doi.org/10.1111/mec.13304.
Puritz JB, Lotterhos KE. Expressed exome capture sequencing: A method for cost-effective exome sequencing for all organisms. Mol Ecol Resour. 2018;18(6):1209-22. https://pubmed.ncbi.nlm.nih.gov/29791785. https://doi.org/10.1111/1755-0998.12905.
Chilamakuri CSR, Lorenz S, Madoui MA, et al. Performance comparison of four exome capture systems for deep sequencing. BMC genomics. 2014;15(1):449. https://pubmed.ncbi.nlm.nih.gov/24912484. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4092227. https://doi.org/10.1186/1471-2164-15-449.
Shendure J, Ji H. Next-generation DNA sequencing. Nat Biotechnol. 2008;26(10):1135-45. https://pubmed.ncbi.nlm.nih.gov/18846087. https://doi.org/10.1038/nbt1486.
Stawski H. Preparing whole genome human mitochondrial DNA libraries for next generation sequencing using Illumina Nextera XT. Western Carolina University; 2013. https://libres.uncg.edu/ir/wcu/f/Stawski2013.pdf.
Bolger AM, Lohse M, Usadel B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114-20. https://pubmed.ncbi.nlm.nih.gov/24695404. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4103590. https://doi.org/10.1093/bioinformatics/btu170.
Li H, Durbin R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics. 2009;25(14):1754-60. https://pubmed.ncbi.nlm.nih.gov/19451168. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2705234. https://doi.org/10.1093/bioinformatics/btp324.
Pirooznia M, Kramer M, Parla J, et al. Validation and assessment of variant calling pipelines for next-generation sequencing. Hum Genomics. 2014;8(1):14. https://pubmed.ncbi.nlm.nih.gov/25078893. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4129436. https://doi.org/10.1186/1479-7364-8-14.
Wang K, Li M, Hakonarson H. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic Acids Res. 2010;38(16):e164. https://pubmed.ncbi.nlm.nih.gov/20601685. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2938201. https://doi.org/10.1093/nar/gkq603.
Prasad RB, Groop L. Genetics of type 2 diabetes—pitfalls and possibilities. Genes (Basel). 2015;6(1):87-123. https://pubmed.ncbi.nlm.nih.gov/25774817. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4377835. https://doi.org/10.3390/genes6010087.
Behjati S, Tarpey PS. What is next generation sequencing?. Arch Dis Child Educ Pract Ed. 2013;98(6):236-8. https://pubmed.ncbi.nlm.nih.gov/23986538. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3841808. https://doi.org/10.1136/archdischild-2013-304340.
Gaulton K, Flannick J, Fuchsberger C. Whole genome and exome sequencing of type 2 diabetes. genetics in diabetes. Gloyn AL, McCarty MI (eds). Genetics in diabetes. Type 2 diabetes and related traits. Front Diabetes. Basel, Karger: Karger Publishers; 2014. https://doi.org/10.1159/000362465.
Bodhini D, Radha V, Deepa R, et al. The G1057D polymorphism of IRS-2 gene and its relationship with obesity in conferring susceptibility to type 2 diabetes in Asian Indians. Int J Obes (Lond). 2007;31(1):97-102. https://pubmed.ncbi.nlm.nih.gov/16652127. https://doi.org/ 10.1038/sj.ijo.0803356.
Shimajiri Y, Sanke T, Furuta H, et al. A missense mutation of Pax4 gene (R121W) is associated with type 2 diabetes in Japanese. Diabetes. 2001;50(12):2864-9. https://pubmed.ncbi.nlm.nih.gov/11723072.
Han X, Xiao J, Ren Q, Tang Y, Yang W, Ji L. Evaluation of variant A45T in NEUROD1/BETA2 for its association with type 2 diabetes mellitus. Endocrine 2013;44(1):99-106. https://pubmed.ncbi.nlm.nih.gov/23203005. https://doi.org/10.1007/s12020-012-9844-3.
Mita M, Miyake K, Zenibayashi M, et al. Association study of the effect of WFS1 polymorphisms on risk of type 2 diabetes in Japanese population. Kobe J Med Sci. 2008;54(4):E192-9. https://pubmed.ncbi.nlm.nih.gov/19258739.
Kifagi C, Makni K, Mnif F, et al. Association of calpain-10 polymorphisms with type 2 diabetes in the Tunisian population. Diabet Metab. 2008;34(3):273-8. https://pubmed.ncbi.nlm.nih.gov/18487065. https://doi.org/10.1016/j.diabet.2008.01.007.
Dupuis J, Langenberg C, Prokopenko I, et al. New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet. 2010;42(2):105-16. https://pubmed.ncbi.nlm.nih.gov/20081858. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3018764. https://doi.org/10.1038/ng.520.
Hattersley AT. Prime suspect: The TCF7L2 gene and type 2 diabetes risk. J Clin Invest. 2007;117(8):2077-9. https://pubmed.ncbi.nlm.nih.gov/17671643. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC1934573. https://doi.org/10.1172/JCI33077.
Kooner JS, Saleheen D, Sim X et al. Genome-wide association study in individuals of South Asian ancestry identifies six new type 2 diabetes susceptibility loci. Nat Genet. 2011;43(10):984-9. https://pubmed.ncbi.nlm.nih.gov/21874001. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3773920. https://doi.org/10.1038/ng.921.
Qi L, Cornelis MC, Kraft P, et al. Genetic variants at 2q24 are associated with susceptibility to type 2 diabetes. Hum Mol Genet. 2010;19(13):2706-15. https://pubmed.ncbi.nlm.nih.gov/20418489. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2883345. https://doi.org/10.1093/hmg/ddq156.
Voight BF, Scott LJ, Steinthorsdottir V, et al. Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet. 2010;42(7):579-89. https://pubmed.ncbi.nlm.nih.gov/20581827. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3080658. https://doi.org/10.1038/ng.609.
Sujjitjoon J, Kooptiwut S, Chongjaroen N, et al. PAX4 R192H and P321H polymorphisms in type 2 diabetes and their functional defects. J Hum Genet. 2016;61(11):943-9. https://pubmed.ncbi.nlm.nih.gov/27334367. https://doi.org/10.1038/jhg.2016.80.
Haghani K, Bakhtiyari S. The study on the relationship between IRS-1 Gly972Arg and IRS-2 Gly1057Asp polymorphisms and type 2 diabetes in the Kurdish ethnic group in West Iran. Genet Test Mol Biomarkers. 2012;16(11):1270-6. https://pubmed.ncbi.nlm.nih.gov/22994406. https://doi.org/10.1089/gtmb.2012.0160.
Lin J, Wang Y, Tang W et al. Insulin receptor substrate-2 (IRS-2) rs1805097 G>A polymorphism is associated with susceptibility to colorectal cancer: A meta-analysis involving 11,234 subjects. Int J Clin Exp Med. 2016;9(7):12639-48.
Willer CJ, Bonnycastle LL, Conneely KN, et al. Screening of 134 single nucleotide polymorphisms (SNPs) previously associated with type 2 diabetes replicates association with 12 SNPs in nine genes. Diabetes 2007;56(1):256-64. https://pubmed.ncbi.nlm.nih.gov/17192490. https://doi.org/ 10.2337/db06-0461.
Palmer, CJ, Bruckner, RJ, Paulo, JA et al. Cdkal1, a type 2 diabetes susceptibility gene, regulates mitochondrial function in adipose tissue. Mol Metab. 2017;6(10):1212-25. https://pubmed.ncbi.nlm.nih.gov/29031721. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5641635. https://doi.org/10.1016/j.molmet.2017.07.013.
Kirchhoff K, Machicao F, Haupt A, et al. Polymorphisms in the TCF7L2, CDKAL1, and SLC30A8 genes are associated with impaired proinsulin conversion. Diabetologia. 2008;51(4):597-601. https://pubmed.ncbi.nlm.nih.gov/18264689. https://doi.org/10.1007/s00125-008-0926-y.
Franks PW, Rolandsson O, Debenham SL, et al. Replication of the association between variants in WFS1 and the risk of type 2 diabetes in European populations. Diabetologia 2008;51(3):458-63. https://pubmed.ncbi.nlm.nih.gov/18040659. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2670195. https://doi.org/10.1007/s00125-007-0887-6.
Sandhu, MS, Weedon, MN, Fawcett, KA et al. Common variants in WFS1 confer risk of type 2 diabetes. Nat Gen. 2007;39(8):951-3. https://pubmed.ncbi.nlm.nih.gov/17603484. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2672152. https://doi.org/10.1038/ng2067.
Fawcett KA, Wheeler E, Morris AP, et al. Detailed investigation of the role of common and low-frequency WFS1 variants in type 2 diabetes risk. Diabetes 2010;59(3):741-6. https://pubmed.ncbi.nlm.nih.gov/20028947. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2828659. https://doi.org/10.2337/db09-0920.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2023 Asif jan, Muhammad Saeed, Zaki ullah, Rani Akbar

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
Journal of the ASEAN Federation of Endocrine Societies is licensed under a Creative Commons Attribution-NonCommercial 4.0 International. (full license at this link: http://creativecommons.org/licenses/by-nc/3.0/legalcode).
To obtain permission to translate/reproduce or download articles or use images FOR COMMERCIAL REUSE/BUSINESS PURPOSES from the Journal of the ASEAN Federation of Endocrine Societies, kindly fill in the Permission Request for Use of Copyrighted Material and return as PDF file to jafes@asia.com or jafes.editor@gmail.com.
A written agreement shall be emailed to the requester should permission be granted.









