Genetic Variants Associated with Poor Responsiveness to Sulfonylureas in Filipinos with Type 2 Diabetes Mellitus
DOI:
https://doi.org/10.15605/jafes.037.S8Keywords:
genetic variants, sulfonylureas, resistance, Filipino, gliclazide, glimepirideAbstract
Introduction. Sulfonylureas (SUs) are commonly used drugs for type 2 diabetes mellitus (T2DM) in the Philippines. This study aimed to associate genetic variants with poor response to gliclazide and glimepiride among Filipinos.
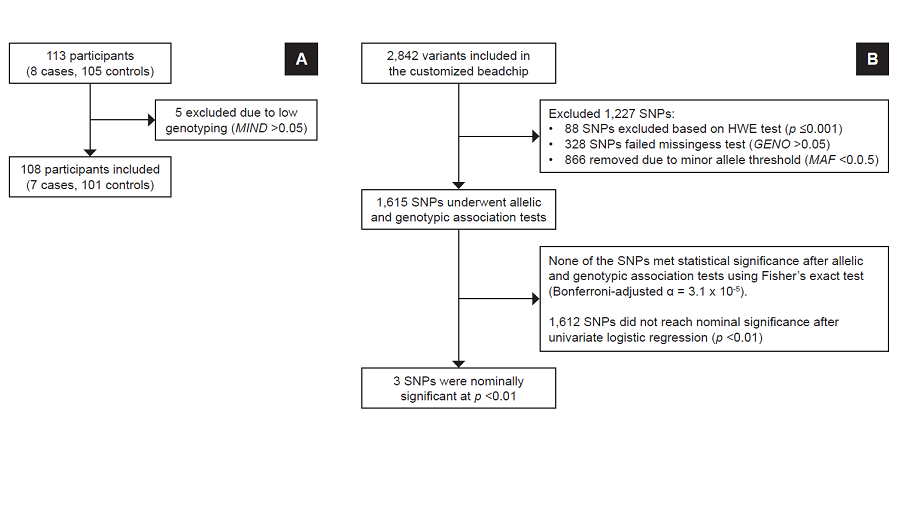
Methodology. Two independent, dichotomous longitudinal substudies enrolled 139 and 113 participants in the gliclazide and glimepiride substudies, respectively. DNA from blood samples underwent customized genotyping for candidate genes using microarray. Allelic and genotypic features and clinical associations were determined using exact statistical methods.
Results. Three months after sulfonylurea monotherapy, 18 (13%) were found to be poorly responsive to gliclazide, while 7 (6%) had poor response to glimepiride. Seven genetic variants were nominally associated (p<0.05) with poor gliclazide response, while three variants were nominally associated with poor glimepiride response. For gliclazide response, carboxypeptidase-associated variants (rs319952 and rs393994 of AGBL4 and rs2229437 of PRCP) had the highest genotypic association; other variants include rs9806699, rs7119, rs6465084 and rs1234315. For glimepiride response, 2 variants were nominally associated: CLCN6-NPPA-MTHFR gene cluster – rs5063 and rs17367504 – and rs2299267 from the PON2 loci.
Conclusion. Genetic variants were found to have a nominal association with sulfonylurea response among Filipinos. These findings can guide for future study directions on pharmacotherapeutic applications for sulfonylurea treatment in this population.
Downloads
References
Sola D, Rossi L, Schianca GP, et al. Sulfonylureas and their use in clinical practice. Arch Med Sci. 2015;11(4):840-8. https://pubmed.ncbi.nlm.nih.gov/26322096. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4548036. https://doi.org/10.5114/aoms.2015.53304.
Jeon JY, Lee SJ, Lee S, et al. Failure of monotherapy in clinical practice in patients with type 2 diabetes: The Korean National Diabetes Program. J Diabetes Investig. 2018;9(5):1144-52. https://pubmed.ncbi.nlm.nih.gov/29328551. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6123024. https://doi.org/10.1111/jdi.12801.
Jimeno C, Sobrepeña L, Mirasol R. DiabCare 2008: Survey on glycaemic control and the status of diabetes care and complications among patients with type 2 diabetes mellitus in the Philippines. Philipp J Intern Med. 2012;50(1):15-22.
Leiter LA, Shestakova MV, Trubitsyna NP, Piletič M, Satman I. Implementing an optimized glucose-lowering strategy with a novel once daily modified release gliclazide formulation. Diabetes Res Clin Pract. 2016;112:50-6. https://pubmed.ncbi.nlm.nih.gov/26653612. https://doi.org/10.1016/j.diabres.2015.11.001.
Garber A, Henry R, Ratner R, et al. Liraglutide versus glimepiride monotherapy for type 2 diabetes (LEAD-3 Mono): A randomised, 52-week, phase III, double-blind, parallel-treatment trial. Lancet. 2009;373(9662):473-81.https://pubmed.ncbi.nlm.nih.gov/18819705. https://doi.org/10.1016/S0140-6736(08)61246-5.
Kim HS, Kim DM, Cha BS, et al. Efficacy of glimepiride/metformin fixed-dose combination vs metformin uptitration in type 2 diabetic patients inadequately controlled on low-dose metformin monotherapy: A randomized, open label, parallel group, multicenter study in Korea. J Diabetes Investig. 2014;5(6):701-8. https://pubmed.ncbi.nlm.nih.gov/25422771. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4234234. https://doi.org/10.1111/jdi.12201.
Kondo Y, Harada N, Hamasaki A, et al. Sitagliptin monotherapy has better effect on insulinogenic index than glimepiride monotherapy in Japanese patients with type 2 diabetes mellitus: A 52-week, multicenter, parallel-group Randomized Controlled trial. Diabetol Metab Syndr. 2016;8:15. https://pubmed.ncbi.nlm.nih.gov/26925169. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4769515. https://doi.org/10.1186/s13098-016-0131-y.
Becker ML, Pearson ER, Tkáč I. Pharmacogenetics of oral antidiabetic drugs. Int J Endocrinol. 2013;2013:686315. https://pubmed.ncbi.nlm.nih.gov/24324494. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3845331. https://doi.org/10.1155/2013/686315.
Huang C, Florez JC. Pharmacogenetics in type 2 diabetes: Potential implications for clinical practice. Genome Med. 2011;3(11):76. https://pubmed.ncbi.nlm.nih.gov/22126607. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3308031. https://doi.org/10.1186/gm292.
Samer CF, Lorenzini KI, Rollason V, Daali Y, Desmeules JA. Applications of CYP450 testing in the clinical setting. Mol Diagn Ther. 2013;17(3):165-84. https://pubmed.ncbi.nlm.nih.gov/23588782. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3663206. https://doi.org/10.1007/s40291-013-0028-5.
Song J, Yang Y, Mauvais-Jarvis F, Wang YP, Niu T. KCNJ11, ABCC8 and TCF7L2 polymorphisms and the response to sulfonylurea treatment in patients with type 2 diabetes: A bioinformatics assessment. BMC Med Genet. 2017;18(1):64. https://pubmed.ncbi.nlm.nih.gov/28587604. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5461698. https://doi.org/10.1186/s12881-017-0422-7.
Holstein A, Hahn M, Körner A, Stumvoll M, Kovacs P. TCF7L2 and therapeutic response to sulfonylureas in patients with type 2 diabetes. BMC Med Genet. 2011;12:30. https://pubmed.ncbi.nlm.nih.gov/21349175. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3051886. https://doi.org/10.1186/1471-2350-12-30.
Tong Y, Lin Y, Zhang Y, et al. Association between TCF7L2 gene polymorphisms and susceptibility to type 2 diabetes mellitus: A large Human Genome Epidemiology (HuGE) review and meta-analysis. BMC Med Genet. 2009;10:15. https://pubmed.ncbi.nlm.nih.gov/19228405. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2653476. https://doi.org/10.1186/1471-2350-10-15.
Suzuki K, Yanagawa T, Shibasaki T, Kaniwa N, Hasegawa R, Tohkin M. Effect of CYP2C9 genetic polymorphisms on the efficacy and pharmacokinetics of glimepiride in subjects with type 2 diabetes. Diabetes Res Clin Pract. 2006;72(2):148-54. https://pubmed.ncbi.nlm.nih.gov/16325295. https://doi.org/10.1016/j.diabres.2005.09.019.
Zhang Y, Si D, Chen X, et al. Influence of CYP2C9 and CYP2C19 genetic polymorphisms on pharmacokinetics of gliclazide MR in Chinese subjects. Br J Clin Pharmacol. 2007;64(1):67-74. https://pubmed.ncbi.nlm.nih.gov/17298483. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2000619. https://doi.org/10.1111/j.1365-2125.2007.02846.x.
Downing KH, Nogales E. Tubulin and microtubule structure. Curr Opin Cell Biol. 1998;10(1):16–22. https://pubmed.ncbi.nlm.nih.gov/9484591. https://doi.org/10.1016/s0955-0674(98)80082-3.
UniProt Consortium. UniProt: The universal protein knowledgebase in 2021. Nucleic Acids Res. 2021;49(D1):D480-9. https://pubmed.ncbi.nlm.nih.gov/33237286. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7778908. https://doi.org/10.1093/nar/gkaa1100.
Thorn CF, Klein TE, Altman RB. PharmGKB: The Pharmacogenomics Knowledge Base. Methods Mol Biol. 2013;1015:311-20. https://pubmed.ncbi.nlm.nih.gov/23824865. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4084821. https://doi.org/10.1007/978-1-62703-435-7_20.
Welter D, MacArthur J, Morales J, et al. The NHGRI GWAS Catalog, a curated resource of SNP-trait associations. Nucleic Acids Res. 2014;42(Database issue):D1001-6. https://pubmed.ncbi.nlm.nih.gov/24316577. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3965119. https://doi.org/10.1093/nar/gkt1229.
Kent WJ, Sugnet CW, Furey TS, et al. The human genome browser at UCSC. Genome Res. 2002;12(6):996-1006. https://pubmed.ncbi.nlm.nih.gov/12045153. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC186604. https://doi.org/10.1101/gr.229102.
Mao J, Ai J, Zhou X, et al. Transcriptomic profiles of peripheral white blood cells in type II diabetes and racial differences in expression profiles. BMC Genomics. 2011;12 Suppl 5(Suppl 5):S12. https://pubmed.ncbi.nlm.nih.gov/22369568. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3287494. https://doi.org/10.1186/1471-2164-12-S5-S12.
Wallingford N, Perroud B, Gao Q, et al. Prolylcarboxypeptidase regulates food intake by inactivating alpha-MSH in rodents. J Clin Invest. 2009;119(8):2291-303. https://pubmed.ncbi.nlm.nih.gov/19620781. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2719925. https://doi.org/10.1172/JCI37209.
Jeong JK, Diano S. Prolyl carboxypeptidase and its inhibitors in metabolism. Trends Endocrinol Metab. 2013;24(2):61-7. https://pubmed.ncbi.nlm.nih.gov/23245768. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3893043. https://doi.org/10.1016/j.tem.2012.11.001.
Zhou C, Garcia-Calvo M, Pinto S, et al. Design and synthesis of prolylcarboxypeptidase (PrCP) inhibitors to validate PrCP as a potential target for obesity. J Med Chem. 2010;53(19):7251-63. https://pubmed.ncbi.nlm.nih.gov/20857914. https://doi.org/10.1021/jm101013m.
Tabrizian T, Hataway F, Murray D, Shariat-Madar Z. Prolylcarboxypeptidase gene expression in the heart and kidney: Effects of obesity and diabetes. Cardiovasc Hematol Agents Med Chem. 2015;13(2):113-23. https://pubmed.ncbi.nlm.nih.gov/26362276. https://doi.org/10.2174/1871525713666150911112916.
[GTExa] Genotype-Tissue Expression. Variant Page – rs2229437. Available in https://gtexportal.org/home/snp/rs2229437. Retrieved on 10 April 2022.
Bai X, Zhang B, Wang P, et al. Effects of SLCO1B1 and GATM gene variants on rosuvastatin-induced myopathy are unrelated to high plasma exposure of rosuvastatin and its metabolites. Acta Pharmacol Sin. 2019;40(4):492-9. https://pubmed.ncbi.nlm.nih.gov/29950617. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC6461793. https://doi.org/10.1038/s41401-018-0013-y.
Liu M, Fan F, Zhang Y, Li J. The association of GATM polymorphism with statin-induced myopathy: A systematic review and meta-analysis. Eur J Clin Pharmacol. 2021;77(3):349-57. https://pubmed.ncbi.nlm.nih.gov/33051696. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7867530. https://doi.org/10.1007/s00228-020-03019-3.
La TM, Yamada H, Seiriki S, et al. Internalization of AMPA-type glutamate receptor in the MIN6 pancreatic β-cell line. Cell Struct Funct. 2020;45(2):121-30. https://pubmed.ncbi.nlm.nih.gov/32581155. https://doi.org/10.1247/csf.20020.
[GTExb] Genotype-Tissue Expression. Variant Page – rs7119. Available in https://gtexportal.org/home/snp/rs7119. Retrieved on 22 April 2022.
Mellado-Gil JM, Fuente-Martín E, Lorenzo PI, et al. The type 2 diabetes associated HMG20A gene is mandatory for islet beta cell functional maturity. Cell Death Dis. 2018;9(3):279. https://pubmed.ncbi.nlm.nih.gov/29449530. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5833347. https://doi.org/10.1038/s41419-018-0272-z.
Colagiuri S, Matthews D, Leiter LA, Chan SP, Sesti G, Marre M. The place of gliclazide MR in the evolving type 2 diabetes landscape: A comparison with other sulfonylureas and newer oral antihyperglycemic agents. Diabetes Res Clin Pract. 2018;143:1-14. https://pubmed.ncbi.nlm.nih.gov/29802958. https://doi.org/10.1016/j.diabres.2018.05.028.
[GTExc] Genotype-Tissue Expression. Variant Page – rs17367504. Available in https://gtexportal.org/home/snp/rs17367504. Retrieved on 22 April 2022.
Del Greco MF, Pattaro C, Luchner A, et al. Genome-wide association analysis and fine mapping of NT-proBNP level provide novel insight into the role of the MTHFR-CLCN6-NPPA-NPPB gene cluster. Hum Mol Genet. 2011;20(8):1660-71. https://pubmed.ncbi.nlm.nih.gov/21273288. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3063986. https://doi.org/10.1093/hmg/ddr035.
Rangaraju A, Krishnan S, Aparna G, Sankaran S, Mannan AU, Rao BH. Genetic variants in post myocardial infarction patients presenting with electrical storm of unstable ventricular tachycardia. Indian Pacing Electrophysiol J.;2018;18(3):91-4. https://pubmed.ncbi.nlm.nih.gov/29396286. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5986548. https://doi.org/10.1016/j.ipej.2018.01.003.
Settin A, El-Baz R, Ismaeel A, Tolba W, Allah WA. Association of ACE and MTHFR genetic polymorphisms with type 2 diabetes mellitus: Susceptibility and complications. J Renin Angiotensin Aldosterone Syst. 2015;16(4):838-43. https://pubmed.ncbi.nlm.nih.gov/24452036. https://doi.org/10.1177/1470320313516172.
Shen Y, Wang Z, Zhou F, Jin R. The influence of MTHFR genetic polymorphisms on methotrexate therapy in pediatric acute lymphoblastic leukemia. Open Life Sci. 2021;16(1):1203-12. https://pubmed.ncbi.nlm.nih.gov/34761111. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8572804. https://doi.org/10.1515/biol-2021-0121.
Wójcicka G, Jamroz-Wiśniewska A, Marciniak A, Łowicka E, Bełtowski J. The differentiating effect of glimepiride and glibenclamide on paraoxonase 1 and platelet-activating factor acetylohydrolase activity. Life Sci. 2010 ;87(3-4):126-32. https://pubmed.ncbi.nlm.nih.gov/20638992. https://doi.org/10.1016/j.lfs.2010.05.018.
Li Q, Tang TT, Jiang F, et al. Polymorphisms of the KCNQ1 gene are associated with the therapeutic responses of sulfonylureas in Chinese patients with type 2 diabetes. Acta Pharmacol Sin. 2017;38(1):80-9. https://pubmed.ncbi.nlm.nih.gov/27694910. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5220536. https://doi.org/10.1038/aps.2016.103.
Naja K, Salami A, El Shamieh S, Fakhoury R. rs622342 in SLC22A1, CYP2C9*2 and CYP2C9*3 and glycemic response in individuals with type 2 diabetes mellitus receiving metformin/sulfonylurea combination therapy: 6-month follow-up study. J Pers Med. 2020;10(2):53. https://pubmed.ncbi.nlm.nih.gov/32575674. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC7354490. https://doi.org/10.3390/jpm10020053.
Lin CH, Lee YS, Huang YY, Hsieh SH, Chen ZS, Tsai CN. Polymorphisms of GLP-1 receptor gene and response to GLP-1 analogue in patients with poorly controlled type 2 diabetes. J Diabetes Res. 2015;2015:176949. https://pubmed.ncbi.nlm.nih.gov/25785276. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4345081. https://doi.org/10.1155/2015/176949.
Didari E, Sarhangi N, Afshari M, Aghaei Meybodi HR, Hasanzad M. A pharmacogenetic pilot study of CYP2C9 common genetic variant and sulfonylureas therapeutic response in type 2 diabetes mellitus patients. J Diabetes Metab Disord. 2021;20(2):1513-9. https://pubmed.ncbi.nlm.nih.gov/34900803. https://www.ncbi.nlm.nih.gov/pmc/articles/PMC8630254 (available on 2022-09-14). https://doi.org/10.1007/s40200-021-00894-0.
Downloads
Published
How to Cite
Issue
Section
License
Copyright (c) 2022 Elizabeth Paz-Pacheco, Jose Nevado Jr., Eva Maria Cutiongco-de la Paz, Gabriel Jasul Jr., Aimee Yvonne Criselle Aman, Elizabeth Laurize Alejandro - Ribaya, Mark David Francisco, Ma. Luz Vicenta Guanzon, May Uyking - Naranjo, Ma. Cecilia Añonuevo - Cruz, Maria Patricia Deanna Maningat, Cristina Jaring, Paulette Nacpil - Dominguez, Aniza Pala-Mohamad, Abigail Uy-Canto, John Paul Quisumbing, Annabelle Marie Lat, Diane Carla Bernardo, Noemie Marie Mansibang, Vincent Sean Ribaya, Karell Jo Angelique Calpito, Julius Patrick Ferrer, Jessica Biwang, Jodelyn Melegrito, Christian Deo Deguit, Carlos Emmanuel Panerio

This work is licensed under a Creative Commons Attribution-NonCommercial 4.0 International License.
The full license is at this link: http://creativecommons.org/licenses/by-nc/3.0/legalcode).
To obtain permission to translate/reproduce or download articles or use images FOR COMMERCIAL REUSE/BUSINESS PURPOSES from the Journal of the ASEAN Federation of Endocrine Societies, kindly fill in the Permission Request for Use of Copyrighted Material and return as PDF file to jafes@asia.com or jafes.editor@gmail.com.
A written agreement shall be emailed to the requester should permission be granted.










